
|
Augustus /
PacBioGMAPBelow I describe the protocol I used to identify new mouse isoforms using single molecule Pacific Bioscience (PacBio) RNA-Seq reads. For clarity these command lines are without parallel execution. However, in reality I split the mouse genome by chromosomes and ran alignments in parallel. The input were circular consensus sequences (ccs), that often constitute near-full length transcripts. The reads were not corrected using other short read data, although I suspect this may improve mapping accuracy. In this example, the input genome is softmasked, i.e. in 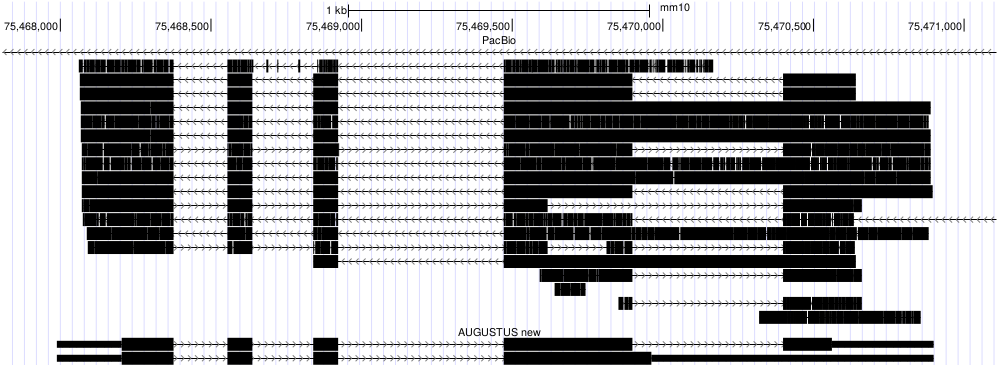 PacBio ccs reads aligned with GMAP and AUGUSTUS predictions. 1. Align the reads with GMAP (Thomas Wu)mkdir gindex gmap_build -D gindex/ -d mm10 mouse_genome.fa gmap -D gindex/ -d mm10 pacbio_rnaseq.fa --min-intronlength=30 --intronlength=500000 --trimendexons=20 -f 1 -n 0 > gmap.psl The option 2. Make hints for AUGUSTUS from alignmentscat gmap.psl | sort -n -k 16,16 | sort -s -k 14,14 | perl -ne '@f=split; print if ($f[0]>=100)' | blat2hints.pl --source=PB --nomult --ep_cutoff=20 --in=/dev/stdin --out=gmap.pacbio.hints.gff In this example, I threw away alignments with less than 100 matches, just because the aim was to find new isoforms with some confidence. When you have multiple libraries, I recommend to align them separately and keep hints separately. You can concatenate these hints later with 3. Predict genes genome-wide with AUGUSTUSaugustus --species=human --alternatives-from-evidence=1 --UTR=on --extrinsicCfgFile=extrinsic.M.RM.PB.cfg --hintsfile=hints.gff --softmasking=1 mouse_genome.fa > augustus.gff The file Remarks for parallel execution: If you split the genome into many chunks, for example to run it on a cluster, it is advisable to split the |