
GSNAPIncorporating Illumina RNAseq into AUGUSTUS with GSNAPThis document describes a method for structurally annotating a genome based on deep sequencing of a transcriptome, RNA-Seq. The guide below was adapted from a description of the method we initially developed for and applied in the RNA-Seq based Genome Annotation Assessment Project (rGASP) that can be found at http://bioinf.uni-greifswald.de/augustus/binaries/readme.rnaseq.html. For rGASP, Mario used BLAT. Using GSNAP, the general approach is to generate intron hints for Augustus from the RNA-Seq, which can be used together with hints from other sources if available (like from an existing gene models, ESTs, protein or genomic conservation, MS/MS). You may adjust the below procedure for your needs, for example, if you choose to first assemble RNA-Seq reads to contigs, you may just just treat them as other larger transcript fragments such as ESTs and use BLAT/GMAP+blat2hints.pl to generate the hints. Currently, this documentation only comprises the generation of intronhints from RNAseq data with GSNAP. Intronhints are for most eukaryotic species the most important hints that can be generated from RNAseq data. According to our experience, intronhints also produce good gene predictions if no UTR model is available and UTRs are rarely spliced, that means intronhints are suitable for most genome annotation projects. Exonparthints are in many cases a valuable addition if a good UTR model exists. The generation of exonparthints may be added to this tutorial at a later point in time. You may incorporate RNAseq data with GSNAP in a single-step or in a two-step approach. We call the two-step approach 'iterative mapping'. Single-step means that you map RNAseq data against the genome, create hints from the alignments, predict genes with AUGUSTUS with these genes, and you're done. The optional second step may increase the number of reads aligned to a splice site and may therefore increase gene prediction accuracy. For the second step, you need to generate an exon-exon junction file that you provide to GSNAP as a database. After this second round of alignment, you map the alignments back to genome level, create new hints and run AUGUSTUS with the new hints, again. We recommend the two-step method. 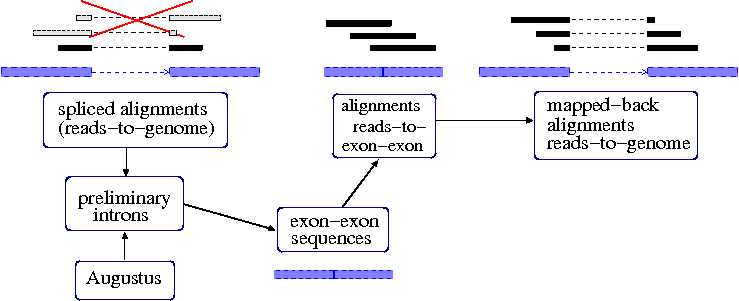 Illustration of iterative mapping Software requirements (make sure to use exactly those versions!): This protocol was tested with the following versions:
Please read the installation instructions of each toolkit carefully and follow their advice! 1. Genome preparationWe recommend that you repeat mask the genome prior mapping RNAseq data to the genome. AUGUSTUS should later be run on the unmasked genome, though. (You should rather include nonexonpart hints generated from repeat regions than running AUGUSTUS on the repeat masked genome.) 2. Read preparation2.1. Read pair namingReads should be in fastq format. If you use paired end reads the two mates need to be in separate files. Furthermore, they need to be in the same order in each file. rnaseq_read1.fastq: @HWUSI-EAS1671_0001:5:1:1022:10290 NCCACTTCTACGATGCCATGGATGGGCAGATACAGGGCAGCGTGGAGCAGGCAGGCCAAGGNNNGGNNAAGGAAG +HWUSI-EAS1671_0001:5:1:1022:10290 ########################################################################### @HWUSI-EAS1671_0001:5:1:1022:10291 NCAAGATCCAAGACAAGGAAGGTATCCCACCAGACCAACAAAGATTGATCTTTGCTGGAAAACAACTTGAAGACG +HWUSI-EAS1671_0001:5:1:1022:10291 ########################################################################### rnaseq_read2.fastq: @HWUSI-EAS1671_0001:5:1:1022:10290 NGCAGCTNNNNNAGGACTNNNNNNNNNNGCTNNNNCNNGNNCCTCCTCTGGAGCAAAACATGACCGGCGTTGGGG +HWUSI-EAS1671_0001:5:1:1022:10290 ########################################################################### @HWUSI-EAS1671_0001:5:1:1022:10291 CGTTTTTCCAGTCAAAGTCTTCACGAAAATTTGCATGCCACCACGGAGACGCAACACCAAGTGGAGGGTGGATTC +HWUSI-EAS1671_0001:5:1:1022:10291 ########################################################################### 2.2. Quality trimmingIn our experiments, we obeserved a decrease in gene prediction accuracy if the reads were quality trimmed prior alignment (we tested accepted error rates of 1% and 5%). Quality trimming does reduce the runtime of GSNAP, though. It's a tradeoff. We recommend to work with untrimmed fastq files if you can afford the computational time. 3. Aligning reads with GSNAP (step 1)You first need to build an index file for your genome: gmap_build -d genome_masked genome_masked.fa Afterwards, align the RNAseq data to the genome: gsnap --nofails --novelsplicing=1 --format=sam -d genome_masked rnaseq.fastq > rnaseq.sam for single read libraries and gsnap --nofails --novelsplicing=1 --format=sam -d genome_masked rnaseq_read1.fastq rnaseq_read2.fastq > rnaseq.sam for paired read libraries. 4. Filtering raw alignments (step 1)samtools view -bS -o rnaseq.bam rnaseq.sam samtools sort rnaseq.bam rnaseq.s samtools sort -n rnaseq.s.bam rnaseq.ss filterBam --uniq --paired --pairwiseAlignment --in rnaseq.ss.bam --out rnaseq.f.bam # remember that you only need to use the flags --paired and --pairwiseAlignment if you actually have paired data! samtools sort rnaseq.f.bam rnaseq.fs # only do this if you have paired data! # later on, we'll need the bam header, therefore extract and store the header: samtools view -H rnaseq.fs.bam > header.txt The last samtools sort command can be omitted if you don't filter for pairedness! 5. Creating intron hints (step1)bam2hints --intronsonly --in=rnaseq.fs.bam --out=hints.gff # in case of paired data # OR bam2hints --intronsonly --in=rnaseq.f.bam --out=hints.gff # in case of unpaired data, when you skipped the sorting step 6. Run Augustus (step 1)6.1. Set hint parametersAdjust the file
[SOURCES]
M RM E W
[GENERAL]
a ss 1 1 0.1 M 1 1e+100 RM 1 1 E 1 1 W 1 1
dss 1 1 0.1 M 1 1e+100 RM 1 1 E 1 1 W 1 1
exonpart 1 .992 .985 M 1 1e+100 RM 1 1 E 1 1 W 1 1.02
intron 1 .34 M 1 1e+100 RM 1 1 E 1 1e6 W 1 1
CDSpart 1 1 .985 M 1 1e+100 RM 1 1 E 1 1 W 1 1
UTRpart 1 1 .985 M 1 1e+100 RM 1 1 E 1 1 W 1 1
nonexonpart 1 1 M 1 1e+100 RM 1 1.15 E 1 1 W 1 1
[Please note that exonpart, CDSpart and UTRpart hints were not produced, here. This feature might be added to the documentation, later.] The exonpart malus of .992 means a weak penalty factor for every predicted exonic base that is not supported by any exonpart hints. The exonpart bonus for hints of source W of 1.02 mean that gene structures get this bonus factor for every exonpart hint of multiplicity 1 that is completely included in an exon. Introns that are not supported by any intron hint are penalized by .34, and introns that are supported by RNA-Seq hints are rewarded by a factor of 10000. The 0.985 are local malus factors. The concept of a local malus accountd for different levels of missing information, that becomes more important with with higher general coverage of RNA-Seq. Like the (normal) malus, the local malus also applies to exonic bases that are unsupported by hints. In contract to the normal malus, the local malus only applies to exons, that are well-supported at some region and not supported at another region of the same exon. Above picture illustrates a typical case where the local malus applies. This UTR exon candidate is very unevenly supported by RNA-Seq coverage and therefore likely to be too long. For every base that is not covered by any exonpart hint a local UTRpart malus factor of 0.985 is applied in addition to the normal exonpart malus of 0.992. Adjust the file [Please note that exonpart, CDSpart and UTRpart hints were not produced, here. This feature might be added to the documentation, later.] The exonpart malus of .992 means a weak penalty factor for every predicted exonic base that is not supported by any exonpart hints. The exonpart bonus for hints of source W of 1.005 mean that gene structures get this bonus factor for every exonpart hint of multiplicity 1 that is completely included in an exon. Introns that are not supported by any intron hint are penalized by .34, and introns that are supported by RNA-Seq hints are rewarded by a factor of 100,000. The 0.985 are local malus factors. The concept of a local malus was recently introduced to account for different levels of missing information, that becomes more important with with higher general coverage of RNA-Seq. Like the (normal) malus, the local malus also applies to exonic bases that are unsupported by hints. In contract to the normal malus, the local malus only applies to exons, that are well-supported at some region and not supported at another region of the same exon. Above picture illustrates a typical case where the local malus applies. This UTR exon candidate is very unevenly supported by RNA-Seq coverage and therefore likely to be too long. For every base that is not covered by any exonpart hint a local UTRpart malus factor of 0.985 is applied in addition to the normal exonpart malus of 0.992. Above parameters have to be seen as an example. The optimal values will depend on various parameters, including 1) Amount of RNA-Seq and percentage of genes expressed in the library. The more transcripts are expressed and covered by RNA-Seq the stronger the malus can be. In the extreme (hypothetical) case, all exonic bases are covered with RNA-Seq and unsupported predicted exonic regions can be punished hard. 2) The stringency of filtering. If, for some other reason, you have chosen to include more than one hit of a read or you allow best alignments also when they are ambiguous, then you may get a significant false positive rate of hints and you may have to reduce bonus factors significantly. 3) Alignment method. 6.2. Run Augustus (step 1)If you don't want to do iterative alignment, you run AUGUSTUS with the following command and stop following this tutorial here: augustus --species=yourSpecies --extrinsicCfgFile=extrinsic.cfg --alternatives-from-evidence=true --hintsfile=hints.gff --allow_hinted_splicesites=atac genome.fa > aug.out For iterative alignment, proceed below this box. augustus --species=yourSpecies --extrinsicCfgFile=extrinsic.cfg --alternatives-from-evidence=true --hintsfile=hints.gff --allow_hinted_splicesites=atac --introns=on --genemodel=complete genome.fa > aug1.out In case you generated exonparthints (not shown here), we recommend switching the UTR flag to "on" because RNA-Seq covers UTR as well. With 7. Create an exon-exon junction database (step 2)All sections starting from here document 'iterative alignment step2' for GSNAP. cat aug1.out | tee aug.prelim.gff | grep -P "\tintron\t" > aug1.introns.gff We combine the AUGUSTUS introns with introns that were previously identified by RNAseq mapping and create a unique list of introns: # in case hints.gff also contains other hints than "intron", you need to filter for "intron", first! cat hints.gff aug1.introns.gff | perl -ne '@array = split(/\t/, $_);print "$array[0]:$array[3]-$array[4]\n";'| sort -u > introns.lst Now, we excise the exon-exon-junctions and write them into fasta format: intron2exex.pl --introns=introns.lst --seq=genome.masked.fa --exex=exex.fa --map=map.psl Note that intron2exex.pl by default creates a junction library of 150 nt length, i.e. 75 nt from each adjacent exon are joined. If your RNAseq library has a read length that is substantially longer than 75 bp, you should either adapt intron2exex.pl to your read length, or treat the RNAseq data like ESTs (e.g. in case of 454 data) instead of treating it like described in this tutorial. The resulting file exex.fa looks like this: >exex246182 75-GTAG-75 chr3.:100000692-100002450 GTTTGGCATCATGGGATGGTATTTTAGACTTGCCAGAACAGAACACTATTCACAAAGATTGCCTGCAGTTTATTGACCAGCTTTCAGTGCCAGAGGAGAAGGCAGCAGAATTACTTTTGGATATTGAATCTGTAATTACCTTTTATTGTA The following command builds a gmap database from the exon-exon junction file: gmap_build -d your_species_exex1 exex.fa 8. Aligning reads with GSNAP (step 2)gsnap --format=sam --nofails -d your_species_exex1 rnaseq.fastq > gsnap.2.sam or for paired reads gsnap --format=sam --nofails -d your_species_exex1 rnaseq_read1.fastq rnaseq_read2.fastq > gsnap.2.sam We need to map the local exex-alignments to global genome level: samMap.pl gsnap.2.sam map.psl > gsnap.2.global.sam 9. Join data from step 1 and step 2Each read has been aligned twice now, to the genome and to the exon-exon sequences. From the read-to-genome alignments we only take the unspliced ones (CIGAR does not contain the letter "N"), as the spliced ones are represented in # discard intron containing alignments from the original bam file bamtools filter -in rnaseq.bam -out rnaseq.noN.bam -script operation_N_filter.txt # create a bam file with header from the gsnap.2.global.sam file cat header.txt gsnap.2.global.sam > gsnap.2.global.h.sam samtools view -bS -o gsnap.2.global.h.bam gsnap.2.global.h.sam # join bam files bamtools merge -in gsnap.2.global.h.bam -in rnaseq.noN.bam -out both.bam If bamtools merge does not work for you (there seem to be version specific problems), you can alternatively try using samtools for the same purpose (this suggestion was reported by Philipp Schiffer, thank you!): samtools merge both.bam gsnap.2.global.h.bam rnaseq.noN.bam Sort: samtools sort -n both.bam both.s 10. Filtering raw alignments (step 2)filterBam --uniq --paired --pairwiseAlignment --in both.s.bam --out both.sf.bam # only use the flags --paired and --pairwiseAlignment if you have paired data! 11a. Creating intron hints (step 2)samtools sort both.sf.bam both.ssf # only do this sorting step if you used the paired flag in filterBam bam2hints --intronsonly --in=both.ssf.bam --out=hints.2.gff # for paired data OR bam2hints --intronsonly --in=both.sf.bam --out=hints.2.gff # for unpaired data 11b. Creating exonpart hints (step 2)While intron hints are pretty 'fail safe' for improving gene structure predictions, exonparthints might in fact mess up your gene predictions. Therefore you carefully need to check gene structures, e.g. in a genome browser, if you decided to include exonparthints from RNAseq data! Generally, exonparthint integration seems to work better if you have parameters for the AUGUSTUS UTR model for your species. Otherwise, AUGUSTUS tends to predict wrong, long exons where in fact there should be UTRs predicted. However, if everything goes well, exonparthints from RNAseq data can improve gene structure predictions significantly! If you previously skipped the sorting step bam2wig both.ssf.bam > both.ssf.wig cat both.ssf.wig | wig2hints.pl --width=10 --margin=10 --minthresh=2 --minscore=4 --prune=0.1 --src=W --type=ep \ --UCSC=unstranded.track --radius=4.5 --pri=4 --strand="." > hints.ep.gff # we'll join and move hints so that you can proceed with this tutorial regardless of whether you created exonparthints, or not: cat hints.ep.gff hints.2.gff > hints.tmp mv hints.tmp hints.2.gff 12. Run Augustus (step 2)augustus --species=yourSpecies --extrinsicCfgFile=extrinsic.cfg --alternatives-from-evidence=true --hintsfile=hints.2.gff --allow_hinted_splicesites=atac genome.fa > aug2.out No warranty for completeness or ability to run. No responsibility for links to external web pages. Contact: augustus-web@uni-greifswald.de |